This post is going to be longer and a little more technical than normal; feel free to jump in and out, or just check out some of the photos on your way to the conclusions. Although I may come across as critical and occasionally cynical at times, I’m not picking on anyone just to be a thorn, but rather to promote scientific discussion; I fully encourage you to join the discussion in the comments section. Finally, in the spirit of full disclosure, a portion of my graduate research was funded via the NSF Tree of Life grant behind this paper (although neither myself nor my research contributed to this project in any manner that I’m aware of), and one of my academic advisors is a co-author on the paper.

Robber Fly with Prey - Asilidae - Ecuador
Despite my best efforts here at Biodiversity in Focus, research on flies very rarely makes the mainstream media (besides mosquitoes, malaria and Drosophila of course), so when one of the most important papers on fly evolution was released and started making the science blog circuit, I was excited to see people taking an interest in Dipterology! There was one problem however, which is not limited to the blogosphere and this paper, but has been an increasingly common trend in insect systematics: the blind acceptance and assumption that a new phylogeny is the definitive answer because the researchers used an ever increasing number of genes. One influential blogger, who’s also an evolutionary entomologist, summarized the results of the Diptera tree of life as such:
But they’re solid results, since they’re based on lots of molecular data and the branch positions are well supported. — Jerry A. Coyne, Ph.D
Similarly, the research team who published the tree are encouraging the idea that their results are infallible by labeling their work the “New Periodic Table of Flies”. A bold statement, and one that many taxonomists might be hesitant to make as it implies that they don’t expect future studies to return different relationships, much as the periodic table of chemical elements is not about to change. An analogy like this requires a strong body of evidence to support it, so let’s take a look at what they did and how the Diptera family tree looks!

Pantophthalmus sp. - Pantophthalmidae - Ecuador
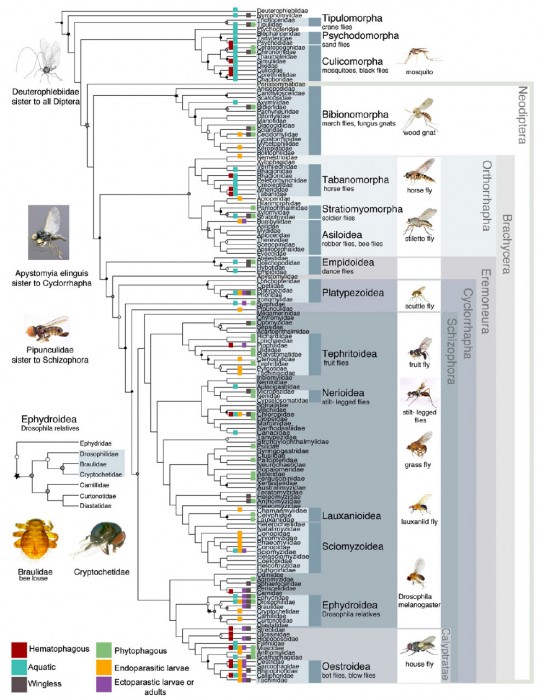
Everything about this paper is large: the 27 member research team represents 18 institutions spread across 3 continents and 6 countries; the 149 families of flies included (there are about 157 families currently recognized); the huge morphological dataset of 371 characters for a subset of the families included; and of course, the use of up to 14 nuclear genes and the entire mitochondrial genome, resulting in a partial dataset of 30,000 basepairs (bp) of DNA data! There are a few important factors regarding the dataset which I should point out however; the massive 30k bp DNA sequences and morphological characters were only obtained for a “relatively” small sampling of fly families (42 “Tier 1″ families) while the remaining 107 families “only” had 5 genes sequenced (~7k bp total). I say “relatively” and “only”, but both of these stipulations are larger than most other papers!
The Tier 1 group was analysed separately using a Bayesian Likelihood analysis (a phylogenetic method which relies on the probability of two species sharing a common ancestor based on mathematical models rather than a direct comparison of similarity as in maximum parsimony) and the results were not actually included in the paper, but relegated as a footnote in the supplementary material (a major downside to PNAS in my opinion).
The results of this massive dataset were actually not too far removed from historical morphological studies. There were a few differences, such as the Bibionomorpha (the group which includes the Bibionidae (March Flies) and the Cecidomyiidae (Gall Midges)) and allies being recovered as the sister to the Brachycera with the Culicomorpha (Culicidae (mosquitoes) and Chironomidae (Midges)) being more distantly related, but overall, pretty similar. Considering this is the portion of the project which tested the “more genes is better” paradigm, the fact that it was left in the seldom-read Supplementary file is rather striking testimony that perhaps more genes isn’t the way of the future.
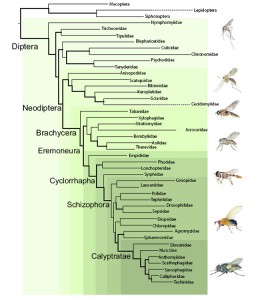
The other general school of thought in insect systematics, which I tend to follow, is the “more taxa is better”, and was ultimately the centerpiece for the paper. Using Maximum Likelihood (another mathematics and probability based method) the research team presented the most comprehensive quantitative analysis of fly relationships:
There is a lot of information packed into that graphic, so we’ll start at the root and work our way through the tree.

Black Fly - Simuliidae - Ecuador
Perhaps the most significant and ecologically satisfying change revolves around the classical “Nematocera”. The families which are largely aquatic as larvae are returned at the root of the tree with the terrestrial “Nematocera” forming a separate clade. It’s easy to imagine the first “fly” developing in water and flies beginning to diversify as they took advantage of different aquatic habitats (i.e. standing water for Culicidae, running water for Simuliidae, etc), and then made the transition to damp terrestrial habitats like fungi (Mycetophilidae, Sciaridae), decomposing vegetation (Bibionidae), or even within living plants (Cecidomyiidae).

Cyphomyia sp. - Stratiomyidae - Bolivia
Within the lower Brachycera, the major shake-up involves the monophyly of the Orthorrhapha and the placement of the Stratiomyomorpha as sister to the Asiloidea rather than the Tabanomorpha. The Orthorrhapha have long been considered to be paraphyletic with regards to the Eremoneura (i.e. one of the lineages of the Orthorrhapha sharing a common ancestor with the Eremoneura), based on the shape of the empodium, an outgrowth of the 5th tarsomere between the tarsal claws. The Stratiomyomorpha and the Tabanomorpha share an empodium which is laterally expanded and flat, while the majority of the remaining Diptera share a narrowed and more bristle-like (filiform) empodium, leading to the Asiloidea being placed as sister to the remaining Diptera by most researchers. By placing the Stratiomyomorpha as sister to the Asiloidea, it implies that the filiform empodium evolved twice, once within the Asiloidea, and again in the Eremoneura. While not impossible for this to have happened, denying the parsimonious assumption of a single evolution will likely be a hard sell to many Dipterists.

Tanypezidae - Ecuador
The majority of the Dipteran families (and species) are found within the Schizophora, and it’s here that I’d argue the tree is least stable. The authors found that this lineage of flies experienced a rapid diversification around 70 million years ago, which has resulted in a huge number of morphologically distinct families, but not enough time for significant changes to accumulate in their DNA. The authors report that 88% of the interfamilial relationships were supported by less than 80 base pairs. 80 base pairs out of 7000. That’s a little over 1% shared DNA between most families in the Schizophora (for the genes examined). Just for comparison’s sake (although not a completely fair comparison I admit), DNA Barcoding proponents usually consider haplotypes which are 2-5% different from one another as indicative of different species. Of course this doesn’t mean that there are no useful results within the Schizophora, as several superfamilies were recovered as monophyletic (including the Nerioidea and Tephritoidea, two groups close to my academic heart), but others are conspicuously missing (Diopsoidea for example).

Snail Killing Fly - Sciomyzidae - Canada
One of the more puzzling superfamilies which is recovered by this study is the Sciomyzoidea, in which the authors have included the Conopidae. I can’t find any reference where the Conopidae are considered members of the Sciomyzoidea, being more commonly referred to as it’s own superfamily (Conopoidea) and placed near the Tephritoidea morphologically or the Diopsoidea in early molecular studies. Carleton University student Joel Gibson recently examined the placement of the Conopidae within the Schizophora in detail. Using a large molecular dataset and representatives of many of the major Schizophoran lineages, he found the Conopidae to be the sister taxon to the remaining Schizophora. Joel also recovered the Conopidae as a single, monophyletic family, compared to the split group recovered in this larger study. The genus Stylogaster has been a point of contention for many years as to whether it actually belongs in the Conopidae. The genus is morphologically distinct and parasitic on cockroaches and crickets which are flushed from the undergrowth by army ant raids, while the remaining Conopidae are generally parasitic on bees and wasps . Joel found Stylogaster to be the basal lineage of the Conopidae, while this study seems to point to a distinct lineage. Regardless of the status of Conopidae, I fail to understand why the authors considered the Sciomyzoidea monophyletic with the Conopoidea smack in the middle of it. Semantics? Sure, but an important distinction in taxonomy nonetheless.

Shore Fly - Ephydridae - Costa Rica
The final groups of flies left to touch on include the Ephydroidea and the Calyptratae, deep within the tree. One of the bigger surprises for this study (and one which the authors made a big deal out of) is the finding that 2 parasitic families (the Braulidae and the Cryptochetidae) which have been difficult to place morphologically, are possibly members of the Drosophilidae. The Drosophilidae are of course best known for Drosophila/Sophophora melanogaster, the poster insect of genetics, but are also one of the most diverse families known, with many different habitats and ecological niches. The Braulidae (bee lice) are morphologically bizarre flies, and as their common name eludes to, are parasites of bee hives, while the Cryptochetidae are endoparasites of scale insects (Hemiptera) as larvae (both families are pictured alongside the tree above). The authors point out that there are species of Drosophilidae which are known to be parasitic in bee hives and of Hemiptera, making the connection between these 3 families very convincing.

Tachinidae - Ecuador
The Ephydroidea was found to be the most likely sister group to the Calyptratae, confirming that the Acalyptratae are a paraphyletic grade of taxa while the Calyptratae are a single monophyletic clade. Although the relationships within the Calyptratae appear rather simple and without much controversy in this study, work published in 2010 examining the relationships within the Oestroidea tells a very different story, with a number of families being found to be paraphyletic. Future work into the relationships of the calyptrate Diptera will likely result in significant changes to the family classification.
Final Comments & Conclusions
The Fly Tree of Life project has achieved a major milestone with this paper, although as evidenced above, we’re a long ways from having all the answers. The researchers have provided evidence that future studies should try and increase the number of species they include, not the number of genes, and while there weren’t many major changes to how we understand fly evolution, there was some strong support for previous hypotheses on higher relationships, and some strange families were confidently placed on the tree. With that in mind, there are a few overall comments I’d like to finish with and which I see as important for future work going forward.

Sphaeroceridae - Costa Rica
Obviously not every species/genus/subfamily of flies can be included in an analysis (yet), but there were a number of families in which the exemplar species used seems odd. Take the Micropezidae for example: 2 species of Calobatinae were included rather than say a Calobatinae and a Taeniapterinae, which are both easily collected in the area surrounding North Carolina State University (the home base for the study). My labmates were equally puzzled by the selections for the Heleomyzidae and Sphaeroceridae. While the authors managed to sample a huge diversity of families and acquire new material suitable for DNA studies for many rare families which are generally hard to find (Ctenostylidae for example), their selection for more common families seems almost like an afterthought. After such a massive study, it’s hard to complain about the taxon sampling, but as more taxa are added to this analysis expect a similar amount of upheaval in family-level relationships.
The other important factor to consider is the paucity of statistical support for most of the nodes within the tree. While there are some well supported clades on the tree, usually between families which had already been identified as having a close affinity or along the back bone of the tree (again lending further credence to accepted divisions), the majority of nodes on the tree did not feature strong support. Even groups which are undoubtedly monophyletic based on morphology, such as the Calyptratae, were found to be poorly supported in this analysis. There’s no reason to accept or reject nodes based solely on support indices, but I would place more confidence in those nodes which are supported.
So is this Fly Tree of Life the “New Periodic Table of Diptera”? No, I don’t think so; there are just too many relationships which aren’t likely to hold up. Is it going to be the most referenced paper in Dipterology for the next 5-10 years? Absolutely. It’s an extremely important paper which sets out a huge number of hypotheses regarding fly relationships which will be tested by every fly taxonomist for years to come. Some relationships will stand up to further testing, some will be refined, and some will be discredited, but there will always be new discoveries made and new relationships proposed. There is plenty left to discover when it comes to flies, and each discovery will impact our understanding of the evolutionary history of flies. Thankfully this is the case, otherwise I’d be out of a job!

Rainieria sp. - Micropezidae - Costa Rica
![]() Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim JW, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, Wheeler BM, Peterson KJ, Pape T, Sinclair BJ, Skevington JH, Blagoderov V, Caravas J, Kutty SN, Schmidt-Ott U, Kampmeier GE, Thompson FC, Grimaldi DA, Beckenbach AT, Courtney GW, Friedrich M, Meier R, & Yeates DK (2011). Episodic radiations in the fly tree of life. Proceedings of the National Academy of Sciences of the United States of America, 108 (14), 5690-5 PMID: 21402926 Open Access
Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim JW, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, Wheeler BM, Peterson KJ, Pape T, Sinclair BJ, Skevington JH, Blagoderov V, Caravas J, Kutty SN, Schmidt-Ott U, Kampmeier GE, Thompson FC, Grimaldi DA, Beckenbach AT, Courtney GW, Friedrich M, Meier R, & Yeates DK (2011). Episodic radiations in the fly tree of life. Proceedings of the National Academy of Sciences of the United States of America, 108 (14), 5690-5 PMID: 21402926 Open Access

Haha first of all thank you for a “real” update, I’ve missed it so…
I have yet to go through the entire paper, but thank you for the detailed breakdown. This is a big step for you dipterists! It is very impressive that they included so many different nuclear loci and the entire mitochondrial genome. I have read a paper recently by Sharanowski et al 2010 on the higher phylogenetic relationships of Hymenoptera using Expressed Sequence Tags (ESTs) instead, and one weakness of this paper was the poor representation of the taxons. These molecular studies often require fresh specimens, making it extremely difficult to include rare specimens.
I will give this a thorough read when I have time and I’m sure I’ll have more to say… I am really digging this whole molecular phylogenetics/phylogenomic business lately hahaha
Thanks for hanging in there for some new posts Miles! I’m hoping to return to a regular blogging schedule soon.
There always seem to be new methods on the horizon as taxonomists find new ways to recover the evolutionary history of their groups! I haven’t heard of the ESTs yet (I’ll have to find that paper it seems) but something this fly paper also presented was using mRNA’s for phylogenetics. I haven’t managed to grasp how it works yet, although I gather that it takes really well preserved specimens to work (like liquid nitrogen storage in the field). Keeping up with the trends is a full time job in itself it seems!
Sharanowski, B. J., Robbertse, B., Walker, J., Voss, S. R., Yoder, S. R., Spatafora, J., Sharkey, M. J. 2010. Expressed sequence tags reveal Proctotrupomorpha (minus Chalcidoidea) as sister to Aculeata (Hymenoptera: Insecta). Molecular Phylogenetics and Evolution 57: 101-112.
If you are interested, she just replaced Dr. Roughley at UofM last year, so hopefully we can hear about her exciting research in the next ESC
Thanks for the reference Miles, I’ll add it to the pile to be read! I see there was another paper published in this month’s Molecular Ecology Resources regarding ESTs:
Lohse, K., B. Sharanowski, M. Blaxter, J. A. Nicholls and G. N. Stone “Developing EPIC markers for chalcidoid Hymenoptera from EST and genomic data.” Molecular Ecology Resources 11(3): 521-529.